A team of researchers has deciphered the genomic blueprint marking the dawn of plant life on land, a pivotal event dating back approximately 550 million years. Led by computational biologist Yanbin Yin from the University of Nebraska–Lincoln, the study unveils the genetic foundations underlying the transition of early terrestrial life forms, including humans.
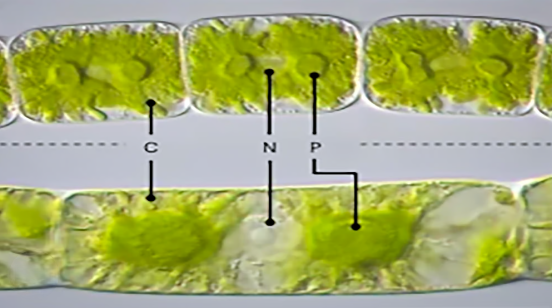
At the heart of the investigation are four genomes belonging to filamentous Zygnematophyceae, the algal relatives of land plants. By analyzing these genomes, the researchers sought to unearth the genetic signatures that paved the way for plants to conquer land.
The emergence of the first land plants, known as Embryophyta, from within the Streptophyta clade around 550 million years ago marks a pivotal moment in evolutionary history. According to the study, the closest living relatives of land plants are the Zygnematophyceae algae, comprising over 4,000 described species distributed across five orders.
The research highlights the adaptability of Zygnematophyceae to terrestrial stressors such as desiccation, ultraviolet radiation, and freezing, suggesting their existence prior to the origin of land plants.
Employing comparative genomics, the researchers aimed to pinpoint genetic innovations characteristic of early land plants. They also investigated the co-expression of genes that evolved in the last common ancestor (LCA) of land plants and Zygnematophyceae.
Leveraging cutting-edge DNA sequencing techniques, the team achieved a complete, chromosome-scale genome assembly for the algae, a notable milestone considering previous sequencing efforts were limited to unicellular Zygnematophyceae.
Yanbin Yin emphasized the evolutionary narrative woven through the study, describing it as a comprehensive account of how early land plants evolved from freshwater algae.
Genome sequencing, the process of unraveling an organism’s complete genetic material, offers invaluable insights into its evolutionary trajectory.
The researchers assembled four multicellular Zygnema strains sourced from algae culture collections at the University of Texas Austin and the University of Göttingen in Germany. Comparative genomic analysis revealed genetic innovations unique to Zygnema, shedding light on its evolutionary trajectory.
The study’s findings, particularly regarding co-expression of genes related to cell wall synthesis and environmental responses, hold implications for various fields including bioenergy, water sustainability, and carbon sequestration.
Jan de Vries, co-corresponding author from the University of Göttingen, underscored the significance of the discovery as a shared resource for the scientific community, offering new avenues for exploration into plant genomics.
Published in the journal Nature Genetics, this study not only enriches our understanding of plant evolution but also underscores the intricate connections between genetic adaptations and environmental responses.